Dec 9, 2024 · 12 min read
Introducing Early Access to the Chan Zuckerberg Initiative’s Platform for AI Cell Models
CZI is building a platform to make biologically relevant AI models and datasets easier to find, evaluate and use. Centralizing and democratizing access to these resources will lower the barriers for scientists to use AI for specific biological tasks and will allow machine learning and AI researchers to rapidly iterate and improve the quality, utility and performance of models, accelerating the pace of scientific research.
Overcoming Barriers to AI-Powered Biological Research
Artificial intelligence has enormous potential to accelerate the pace of scientific discovery. The generation and curation of enormous, biologically rich datasets and advances in machine learning have made it easier to train models to solve the most pressing biological questions of our time, yet several barriers limit the pace and scope of such research.
One is that scientific research remains fragmented. Within industry, generative AI continues to be leveraged for narrow purposes, such as modeling molecular interactions in support of developing targeted therapeutics. Within academia, access to high-powered computing resources can be constrained and costly. Additionally, the way experts evaluate models varies widely, making it hard to establish consistent standards.
Another key challenge is making AI models for biological research easier to develop and use. When it comes to training, fine-tuning and benchmarking models, a major limitation for model developers is the lack of access to curated, standardized datasets that are representative of biology in multiple modalities, scales and population diversities.
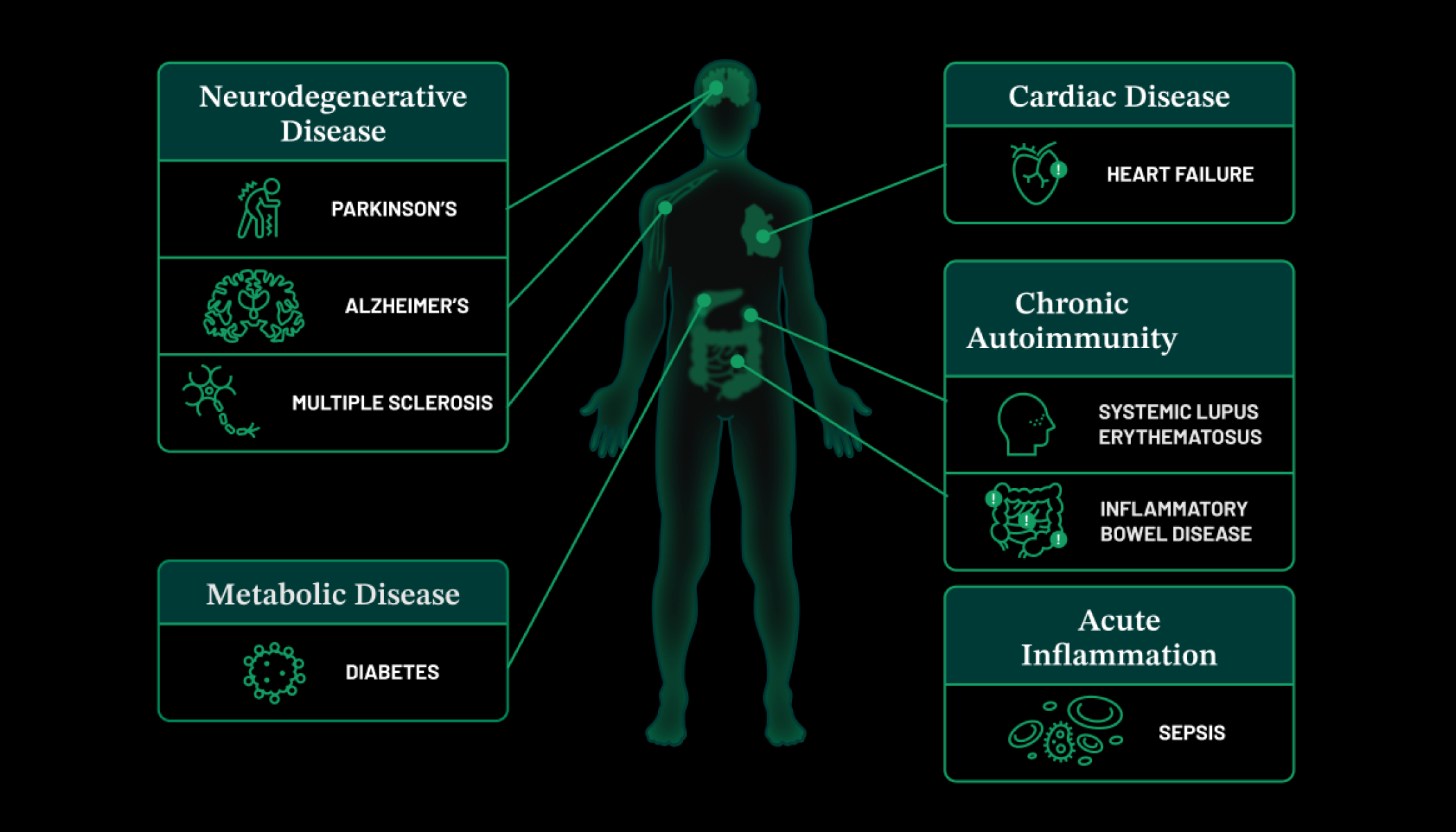
Bringing the power of generative AI to biology at scale will allow researchers to incorporate these technological advances into their work, which will accelerate efforts to cure, prevent, or manage all disease. … AI models could predict how an immune cell responds to an infection, what happens at the cellular level when a child is born with a rare disease, or even how a patient’s body will respond to a new medication. We hope that this collaborative effort will generate new insights about the fundamental characteristics of our cells.
What’s more, model developers are incentivized to publish models quickly without necessarily packaging them in a way that biologists can easily use and reproduce. These challenges are compounded by the lack of extensive coding expertise facing many biologists who may struggle to use models that require complex tasks, such as transforming data for input into a model. By making models easier to use and test for biologists, machine learning experts can gain valuable feedback from the deep subject matter expertise of scientists to improve models for specific biological applications.
As the use of AI for biology rapidly evolves, bringing machine learning researchers and biologists together will help open up new possibilities for collaboration and application across fields — ultimately improving AI’s ability to unlock new insights about human biology.
CZI is releasing a platform that bridges these gaps by providing centralized access to curated models and their underlying datasets — enabling tighter collaborations with the scientific community and technical disciplines. This platform builds on CZI’s efforts to develop software for science and provide access to high-quality, standardized biological data. This includes tools like Chan Zuckerberg CELL by GENE (CZ CELLxGENE), an open-source platform that allows scientists to access, analyze and annotate high-dimensional single-cell data at scale; and the CryoET Data Portal, which provides biologists and developers access to high-quality, standardized, annotated tomograms to retrain or develop annotation models and algorithms.
Earlier this year, we brought together experts to discuss the possibilities of leveraging advances in AI to construct virtual cells — high-fidelity simulations of cells and cellular systems under different conditions that are directly learned from biological data across measurements and scales. These virtual cell models could open up an entirely new era of in-silico experimentation.
Our overarching vision is to leverage advances in generative AI to unlock the mysteries of the cell — a crucial milestone in CZI’s journey toward curing, preventing or managing all diseases by the end of this century.
“Bringing the power of generative AI to biology at scale will allow researchers to incorporate these technological advances into their work, which will accelerate efforts to cure, prevent, or manage all disease,” says CZI Co-Founder and Co-CEO Priscilla Chan. “AI models could predict how an immune cell responds to an infection, what happens at the cellular level when a child is born with a rare disease, or even how a patient’s body will respond to a new medication. We hope that this collaborative effort will generate new insights about the fundamental characteristics of our cells.”
Easier Access to Biologically Relevant AI Models and Data
As the number of AI models for biology rapidly grows, it can be difficult for both machine learning researchers and biologists to easily find, learn about and compare relevant AI models across disparate sources. Unlike generalist platforms, our goal is to build a streamlined platform of curated, state-of-the-art models that have been vetted for biological relevance and usability. Initially, we’re starting with a set of models and datasets in the scientific domains of subcellular imaging and single-cell transcriptomics, built by CZI, our AI residents, collaborators, and the wider scientific community. We will grow the number of available models over time.
Each model has a model card that summarizes important information from multiple sources, including preprints and data repositories, all in one place. This enables researchers to better understand and evaluate key aspects of a model, such as provenance, intended goals and potential applications, training data, hyperparameters, and evaluation and performance metrics. These cards also link to training and evaluation datasets.
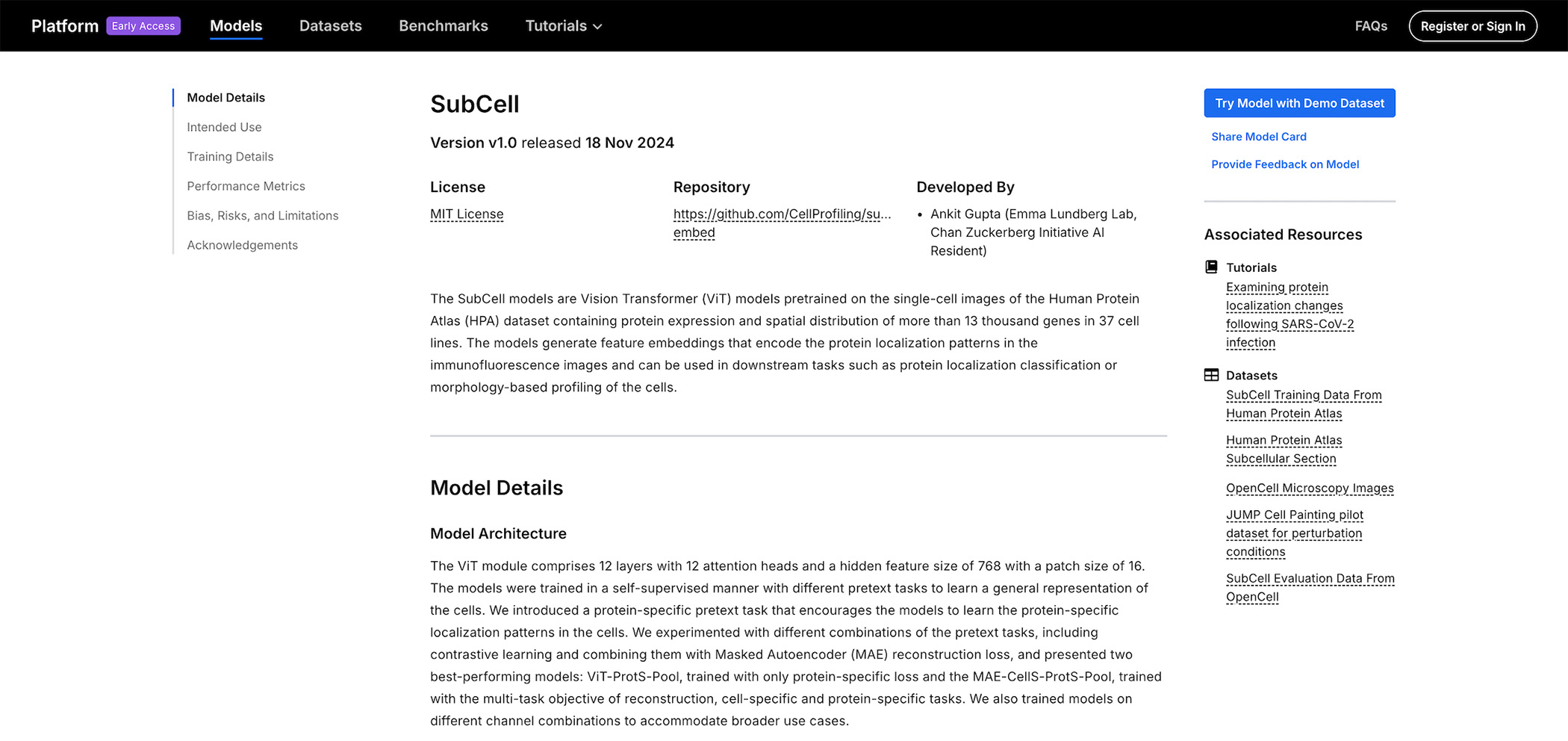
To make these models more accessible to biologists, the platform includes tutorials that show how a model can be applied to a specific biological task. For example, the SubCell model tutorial demonstrates how to use SubCell to examine protein localization changes following SARS-CoV2 infection in microscope images of cells. Biologists can open the tutorial in a Google Colab notebook to easily run the model using the provided demo data. This ensures they can quickly get hands-on experience with the model without worrying about hardware requirements, installation dependencies, or transforming input data to ensure it works with the model. As a next step after running the demo data, the tutorial describes the data processing steps for biologists to run their own fluorescence microscopy image data through the model, so they can learn how they might apply these models to their own research area.
What Researchers Can Do With the Platform
1. Biologists can discover curated cell biology models and datasets and get started applying AI to their research.
Scientists can explore the first set of models on the platform and use the provided model cards and tutorials to more quickly understand if they are relevant to their specific area of research. Initial models developed by CZI and collaborators include scGenePT and SubCell, and those developed by the wider scientific community include scVI, scGPT and CELL-DIFF. Biologists can then start applying these models with notebooks and demo data.
Models that embed imaging data to facilitate image analysis tasks such as protein localization include:
- SubCell: A collection of self-supervised Vision Transformer (ViT) models designed to analyze cell phenotypes and subcellular protein distribution from microscope images.
- CELL-Diff: A diffusion transformer model designed to generate detailed protein localization images from protein sequences (“sequence-to-image”) or output protein sequences based on microscopy images depicting protein localization (“image-to-sequence”).
Models that embed transcriptomic data to analyze patterns of gene expression across cell types and cell states include:
- scGenePT: A collection of single-cell models for perturbation prediction that extends the scGPT foundation model for scRNAseq data by injecting language embeddings at the gene level into the model architecture.
- scGPT: A foundation model designed to integrate and analyze large-scale single-cell multi-omics data using a generative pre-trained transformer (GPT) architecture.
- scVI: A probabilistic deep generative model designed to analyze single-cell RNA sequencing (scRNA-seq) data.
Each model on the platform comes with a notebook to help biologists get the model running quickly with minimal debugging. For instance, researchers can dive into single-cell transcriptomic data analysis with a scVI model trained on 74 million cells from CZ CELLxGENE. They can learn how to leverage the model to integrate and compare their data, find biologically similar cells, and gain deeper insights into cell-type associations. Google Colab notebook and data processing scripts make it easy to get the model running.
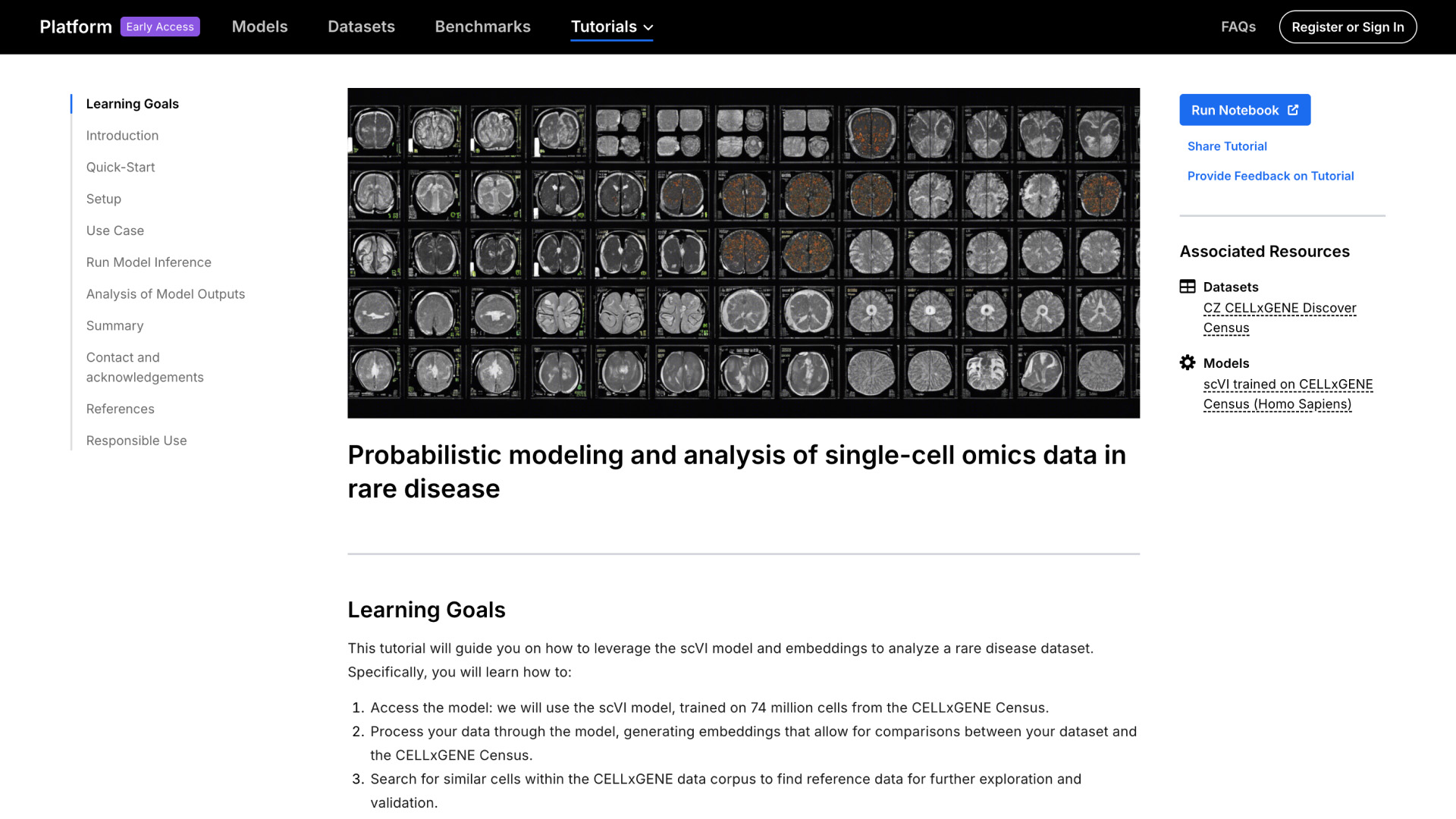
Looking toward the future, our goal is to enable biologists to easily compare, select and use the best model for their specific biological questions without the need for significant computational expertise. We plan to add features, including benchmarks that help researchers compare the models that perform a certain task and choose the best one for their needs, learning resources for biologists on the basics of how to get started using models, and the ability to run models without the need to code.
2. Machine learning researchers can build and improve upon models for biological impact.
The platform provides AI/ML researchers with the ability to quickly and efficiently run models by opening them in a Google Colab notebook. We’ve also linked each of the models to its associated data sources, including processed datasets used to train and evaluate the models. These functionalities unlock a critical data bottleneck, so AI/ML researchers can more efficiently assess underlying datasets and then use those datasets to develop and/or fine-tune models with minimal need for wrangling and transformation.
For example, a machine learning researcher interested in building upon SubCell can access the datasets it was trained and evaluated on, such as:
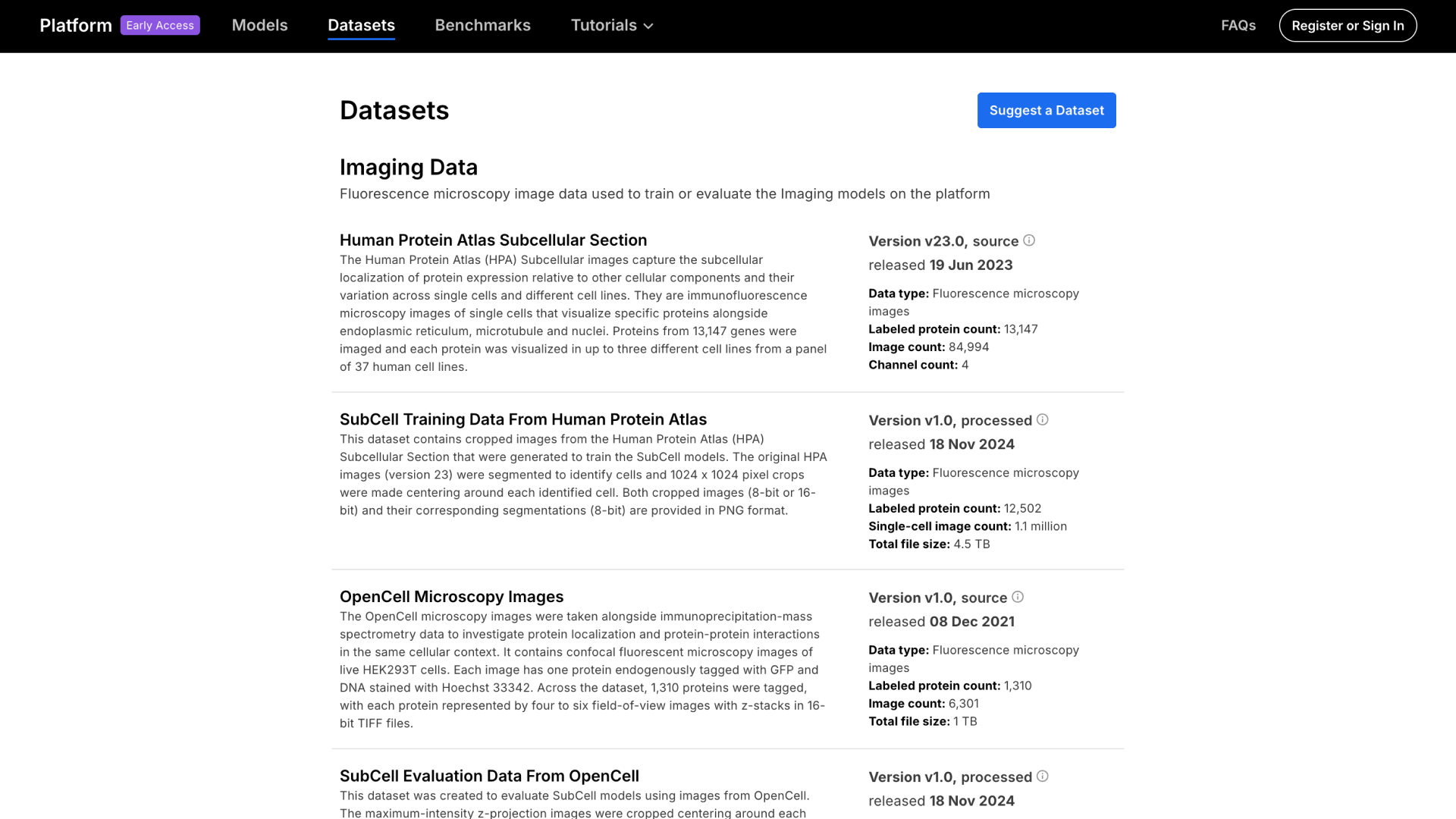
- Human Protein Atlas (HPA) Source Dataset: Images that capture the subcellular localization of proteins relative to other cellular components and their variation across single cells and different cell lines. Proteins from 13,147 genes were imaged.
- HPA for SubCell Processed Dataset: This dataset has been transformed from its source format for model usage that contains cropped images from the HPA subcellular section that were generated to train the SubCell models. The original HPA images were segmented to identify cells and crops were made centering around each identified cell.
- OpenCell Processed Dataset: This dataset was created to evaluate the SubCell model using images from OpenCell. The maximum-intensity z-projection images were cropped, centering around each nucleus.
- JUMP Cell Painting pilot dataset for perturbation conditions Source Dataset: This is a pilot dataset from the JUMP-Cell Painting Consortium that tested both chemical and genetic perturbations in two cell lines U2OS and A549. The perturbations include small molecule drugs, gene knockout and gene overexpression targeting 160 genes and their protein products and two time points were taken.
Additionally, the platform links to another CZI-built tool that accelerates model development, the CZ CELLxGENE Discover Census. This is a large-scale, scRNA-seq resource that integrates data from over 33 million human cells across various tissues, conditions and disease states from human and mouse datasets.
CZ CELLxGENE Census offers API access to the world’s largest standardized single-cell data resource. The API is designed to enable scalable machine learning workflows through the integration of PyTorch data loaders that ensure:
- Efficient data access. Researchers can stream data in chunks via the API, avoiding the need to load the entire dataset into memory.
- Optimized model training. Built-in data loaders handle shuffling and randomization, preventing biases from the dataset’s structured organization and ensuring effective training.
These are only a few of the datasets available on the platform. You can view all available datasets by visiting the dataset collection page.
ML researchers can suggest their own models and datasets for consideration to be included on the platform via a submission form, which will allow them to share their work with a broader audience and receive valuable feedback from the scientific community. CZI is currently prioritizing models that support imaging and transcriptomic research, but we’re also interested in models in other research domains.
We will continue to add models created by both CZI and the community, as well as new features, including benchmarking for comparison of biological models, more mechanisms for feedback from biologists on how to improve models, and API access to query and find relevant training and validation data.
Looking to the Future
This is just the start of our journey towards building virtual cell models that are capable of predicting the behavior of cells and cell systems. Feedback from machine learning experts and biologists will be crucial to training models and providing resources to the community that will one day support the entire biological research lifecycle. We will continue to openly share resources for modeling, evaluating, and analysis of cellular data with the scientific community on this platform.
We are launching a first-of-its-kind request for applications for researchers to build large-scale AI/ML models using compute power on CZI’s high-performance computing cluster to power new approaches to biological discovery. CZI is the only philanthropic organization to fund and build one of the largest computing systems dedicated to nonprofit life sciences research in the world.
Science builds on science, so our teams will collaborate closely with researchers in the field to evolve these initial models — paving the way for a future where scientists can curate virtual cells for any and all cell types across tissues, individuals or conditions.
If you’re interested in using AI for biological impact, please visit the platform and provide feedback.