Cell Science
The CZI Cell Science program (formerly the Single-Cell Biology program) supports and enables the generation and application of technologies for the engineering and research of complex cellular systems. Despite an ever-increasing knowledge of cell types and states, the vast majority of primary cells remain remarkably difficult to culture and experiment with. The Cell Science program seeks to change this by building on their established interdisciplinary collaborations to derive molecular definitions of cells and dig deeper into their biology. At its core, the program is focused on scalable technology that enables deeper insight into cells in health and disease states via experimentation and modeling.
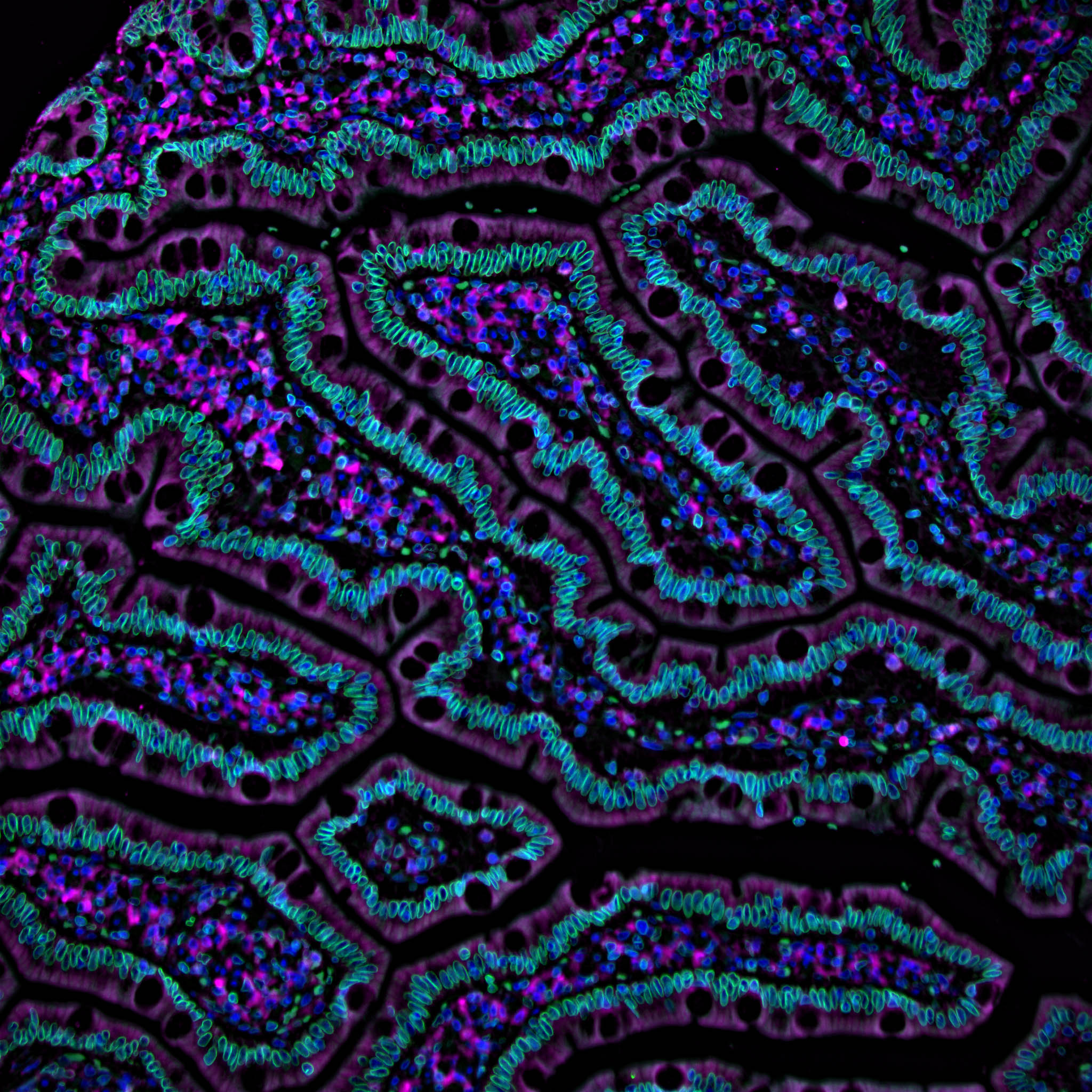
The Cell Science program has funded the following RFAs:
- Measuring Metabolism Across Scales, $8.6 million
- Single-Cell Biology Data Insights (Cycle 1, $5.2 million and Cycle 2, $4.9 million)
- Patient-Partnered Single-Cell Analysis of Rare Pediatric Disease, $10 million
- Ancestry Networks for the Human Cell Atlas, $27.9 million
- Pediatric Networks for the Human Cell Atlas, $34.2 million
- Single-Cell Analysis of Inflammation, $14 million
- Seed Networks for the Human Cell Atlas, $67.7 million
- Collaborative Computational Tools for a Human Cell Atlas, $14.8 million
- Pilot Projects for a Human Cell Atlas, $14.9 million
Single-Cell Technology
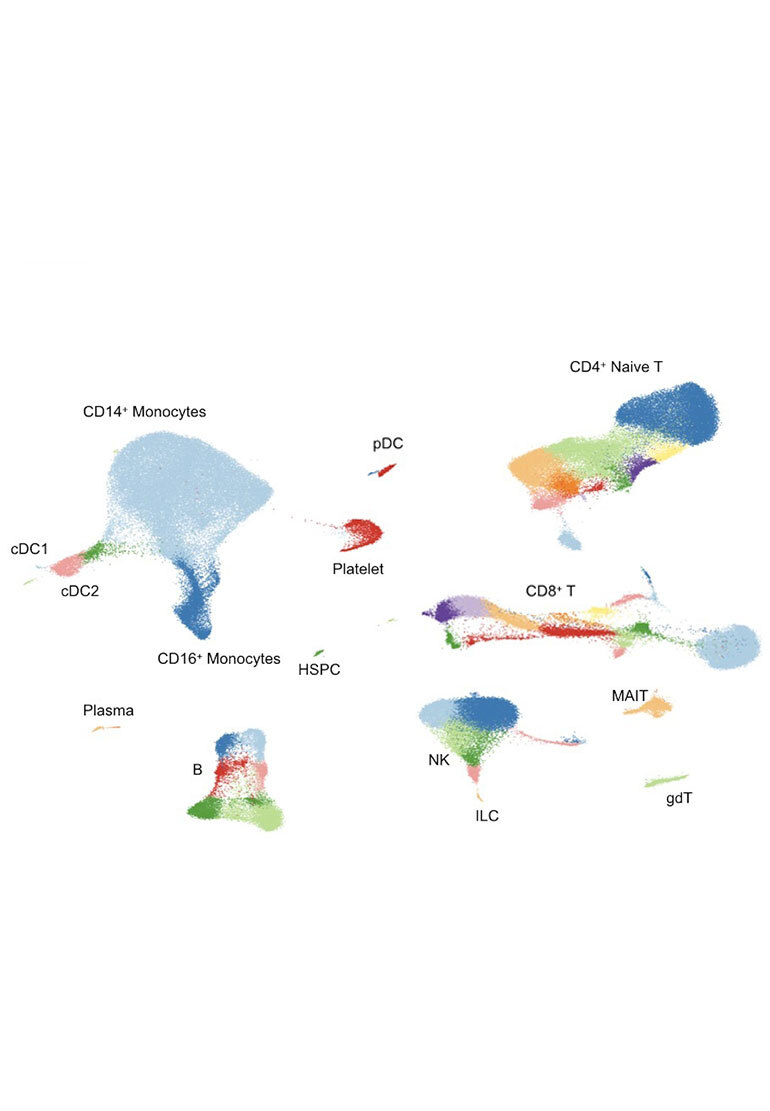
We build tools to support and accelerate the work of biologists who are uncovering new insights about healthy and diseased cells using single-cell methods. Our platform, Chan Zuckerberg CELL by GENE (CZ CELLxGENE), allows scientists to access, analyze, and annotate high-dimensional single-cell data at scale, so they can inform their experiments or quickly surface new insights that could lead to discoveries in treating disease. Learn more at czcellxgene.org and read our pre-print.

CZ CELL×GENE Discover
CZ CELLxGENE Discover provides scientists with fast, easy access to the largest collection of standardized single-cell data — 50+ million cells and growing. Scientists can access the data in different ways based on their research needs:
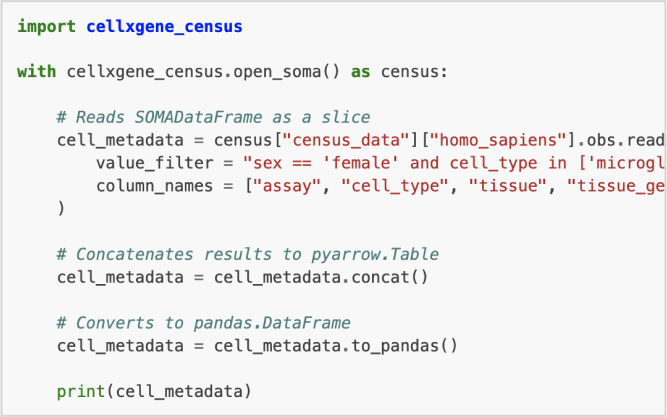
Census
Access any custom slice of standardized cell data available on CZ CELLxGENE Discover via R and Python.
Learn More
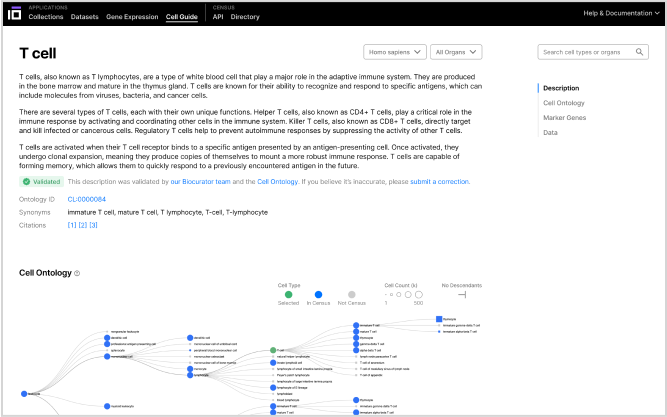
CellGuide
Explore an interactive encyclopedia of 700+ cell types that provides detailed definitions, marker genes, lineage, and relevant datasets in one place.
Learn More
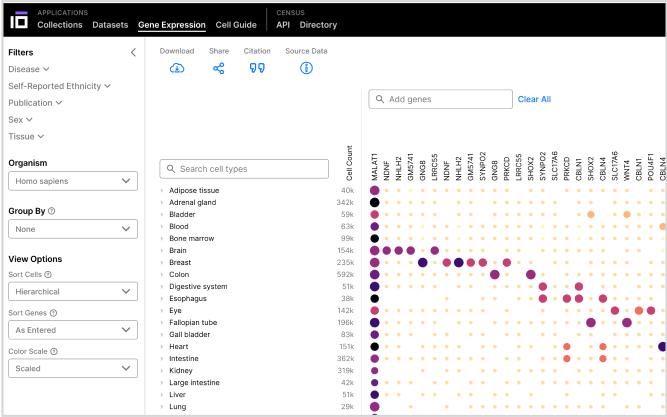
Gene Expression
Uncover expression levels of any gene across datasets of cell types and tissues with advanced visualization tools.
Learn More

CZ CELL×GENE Annotate
CZ CELLxGENE Annotate is a self-hosting interactive data explorer for single-cell datasets. Our desktop tool is a powerful interface that allows you to annotate, analyze, and explore your own single-cell data. Learn more here.
Cellular Measurement
Advancements in technology offer an unprecedented potential to profile and visualize cells, RNA, proteins, and metabolites. This work aims to generate comprehensive data references spanning modalities, ancestries, and developmental stages that can be used to rapidly understand the molecular mechanisms that define cells.
Prediction Engines
There remain many opportunities to use and reuse single-cell data to more deeply interrogate cellular mechanisms and disease. This work aims to equip leading-edge computational scientists with large data resources and unlock untapped power to describe cellular function.
Perturbation and Validation
Perturbation and manipulation of complex cellular systems are critical to extending and validating inferences made by current cell atlases. This work focuses on the development of new technologies that enable scientists to perturb cells and systems at scale across organisms, modalities, and diseases.
Cell Engineering
This work moves beyond functional interrogation and extends into engineering cell identity, fate, or behavior using synthetic biology, with the goal of validating our molecular knowledge of cells.
Funding Opportunities
News & Stories
Interested in learning more about our work in Cell Science? Get the latest information from the links below.
 Contact the Cell Science team
Contact the Cell Science team
To contact the Cell Science team, please complete this form.

Thank you for contacting the Cell Science team!
Your email has been sent.

An error occurred contacting the Cell Science team.
Please try again.
Join Our Science Mailing List
Be the first to know about new funding opportunities.

Join Our Team
Explore our job openings.
Explore Our Grant Resources
Read our Request for Applications.